Something appears amiss with TrEMBL, millions of sequences are “missing”. Where did they go?
At the end of last month, to build a database of bacterial sequences with known hydrolase activity1, I extracted around 2.9 million sequences from UniProtKB/TrEMBL; a popular database which contains sequences that have been automatically annotated and are awaiting manual curation for graduation to the UniProtKB/SwissProt database. It’s important to note that as these annotations have not yet been reviewed they may be less accurate, but it is this same lack of review that allows the database to be so large — making it a useful first port of call when trying to classify your own sequences. Typically we handle the potential for less accurate results by using a more stringent quality threshold (than we would for a manually curated database such as SwissProt, or one we have created in confidence) when filtering alignment hits from software such as BLAST.
It’s good to keep databases up-to-date and so I ran the same query2 against TrEMBL with a view to re-download the resulting FASTA, only to find just shy of 1 million results had been returned — just over a third of the original query a month ago. Wat?
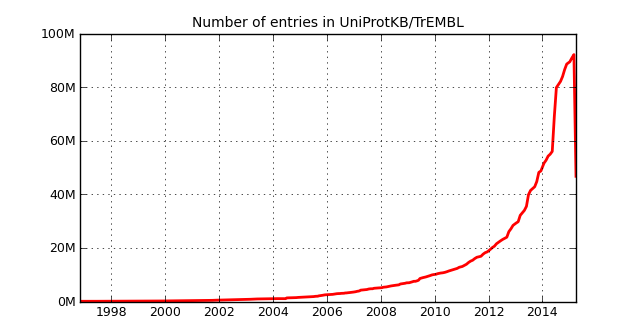
TrEMBL was most recently updated at the start of April3 and the current release notes include the graph below.
Indeed it appears that half of TrEMBL is missing? After my initial panic state, I presumed this must have been a database spring clean to remove similar looking sequences and digging around the FTP repository, my hunch was confirmed in an additional news file:
The UniProt Knowledgebase (UniProtKB) has witnessed an exponential growth in the last few years with a two-fold increase in the number of entries in 2014. This follows the vastly increased submission of multiple genomes for the same or closely related organisms. This increase has been accompanied by a high level of redundancy in UniProtKB/TrEMBL and many sequences are over-represented in the database. […] …we have developed a procedure to identify highly redundant proteomes within species groups using a combination of manual and automatic methods. We have applied this procedure to bacterial proteomes (which constituted 81% of UniProtKB/TrEMBL in release 2015\_03) and sequences corresponding to redundant proteomes (47 million entries) have been removed from UniProtKB. […] From now on, we will no longer create new UniProtKB/TrEMBL records for proteomes identified as redundant.
Personally I would have liked to see this sort of major announcement (and actually a bit more information on “the procedure“) in the release notes4 rather than as an aside stored in an HTML file that I wouldn’t open in my terminal. Though it is amusing that the removal of 47 million entries still wasn’t enough of a story to make it the “Headline” piece of news for the release!
Mystery solved. At least my BLAST jobs have less to hit against now5?
@samstudio8 Are you referring to reduced redundancy in @uniprot ? Here’s some info about it in their Help pages: http://t.co/s8zhqztWpQ
— EMBL-EBI (@emblebi) April 27, 2015
tl;dr
- TrEMBL v2015_04 features 47 million less sequences than v2015_03, to reduce unnecessary redundancy.
|
1 2 3 4 5 6 7 |
[msn@bert databases]$ python3 ~/scripts/summary_stat_fasta.py uniprot-2015_03_27-ec3-tax2_bacteria.fasta #NO 2,900,509 MAX 12,374 MIN 8 AVG 362.95 #NT 1,052,747,787 |
-
Sequences with an EC Number (Enzyme Classification) of
3.*with taxonomic classBacteria [2]. ↩ -
The
reviewed:nosearch query limits results from the UniProtKB to just entries found in TrEMBL. ↩ - And I figured this would make a pretty poor April Fool’s joke. ↩
- A large neon sign wouldn’t have gone amiss either. ↩
- I imagine our sys-admin will be pleased to have some scratch space back too. ↩