Happy New Year!
The guilt of not writing has reached a level where I feel sufficiently obligated to draft a post. You’ll likely notice from the upcoming contents that I am still a PhD student, despite a previous, more optimistic version of myself writing that 2016 would be my final Christmas as a PhD candidate.
Much has happened since my previous Status Report, and I’m sure much of it will spin-off to form several posts of their own, eventually. For the sake of brevity, I’ll give a high level overview.
I’m supposed to be writing a thesis anyway.
Previously on…
We last parted ways with a double–bill status report lamenting the troubles of generating suitable test data for my metagenomic haplotype recovery algorithm, and documenting the ups-and-downs-and-ups-again of analysing one of the synthetic data sets for my pre-print. In particular, I was on a quest to respond to our reviewer’s desire for more realistic data: real reads.
Gretel: Now with real reads!
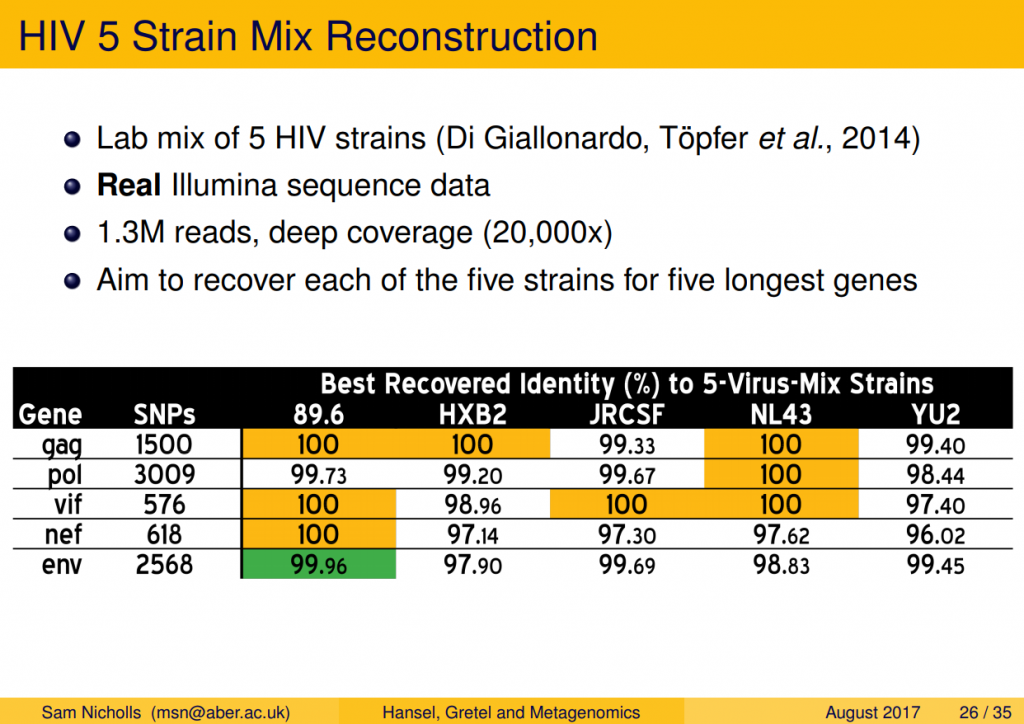
Part Two of my previous report alluded to a Part Three that I never got around to finishing, on the creation and analysis of a test data set consisting of real reads. This was a major concern of the reviewers who gave feedback on our initial pre-print. Without getting into too much detail (I’m sure there’s time for that); I found a suitable data set consisting of real sequence reads from a lab-mix of five HIV strains, used to benchmark algorithms in the related problem of viral-quasispecies reconstruction. After fixing a small bug, and implementing deletion handling, it turns out we do well on this difficult problem. Very well.
In the same fashion as our synthetic DHFR metahaplome, this HIV data set provided five known haplotypes, representing five different HIV-1 strains. Importantly, we were also provided with real Illumina short-reads from a sequencing run containing a mix of the five known strains. This was our holy grail, finally: a benchmark with sequence reads and a set of known haplotypes. Gretel is capable of recovering long, highly variable genes with 100% accuracy. My favourite result is a recovery of env — the ridiculously hyper-variable envelope gene that encodes the HIV-1 virus’ protein shell — with Gretel correctly recovering all but one of 2,568 positions. Not bad.
A new pre-print
Armed with real-reads, and improved results for our original DHFR test data (thanks to some fiddling with bowtie2), we released a new pre-print. The manuscript was a substantial improvement over its predecessor, which meant it was all the more disappointing to be rejected from five different journals. But, more on this misery at another time.
My PhD has sunk to the eating a sad McDonald's in a car park in the dark stage
— Sam Nicholls (@samstudio8) February 17, 2017
mfw reading my paper reviews pic.twitter.com/PWOlUVVWBZ
— Sam Nicholls (@samstudio8) June 12, 2017
Despite our best efforts to address the previous concerns, new reviewers felt that our data sets were still not a good representation of the problem-at-hand: “Where is the metagenome?”. It felt like the goal-posts had moved, suddenly real reads were not enough. But it’s both a frustrating and fair response, work should be empirically validated, but there are no metagenomic data sets with both a set of sequence reads, and known haplotypes. So, it was time to make one.
I’m a real scientist now…
And so, I embarked upon what would become the most exciting and frustrating adventure of my PhD. My first experiences of the lab as a computational biologist is a post sat in draft, but suffice to say that the learning curve was steep. I’ve discovered that there are many different types of water and that they all look the same, that 1ml is a gigantic volume, that you’ll lose your fingerprints if you touch a metal drawer inside a -80C freezer, and that contrary to what I might have thought before, transferring tiny volumes of colourless liquids between tiny tubes without fucking up a single thing, takes a lot of time, effort and skill. I have a new appreciation for the intricate and stochastic nature of lab work, and I understand what it’s like for someone to “borrow” a reagent that you spent hours of your time to make from scratch. And finally, I had a legitimate reason to wear an ill-fitting lab coat that I purchased in my first year (2010), to look cool at computer science socials.
Video of @samstudio8 setting up a PCR – MICROBIOLOGY pic.twitter.com/45cLzz5u65
— Samantha Pendleton (@sap218) September 19, 2017
With this new-found skill-tree to work on, I felt like I was becoming a proper interdisciplinary scientist, but this comes at a cost. Context switching isn’t cheap, and I was reminded of my undergraduate days where I juggled mathematics, statistics and computing to earn my joint honours degree. I had more lectures, more assignments and more exams than my peers, but this was and still is the cost of my decision to become an interdisciplinary scientist.
my wet-dry lab work balance pic.twitter.com/6BeeGSUBsG
— Sam Nicholls (@samstudio8) July 25, 2017
And it was often difficult to find much sympathy from either side of the venn diagram…
My new lab friends wonder why I "take the day off" to push buttons, yet fellow bioinfos are confused it takes a morning to "pipette shit"
— Sam Nicholls (@samstudio8) July 10, 2017
..and science can be awful
I’ve suffered many frustrations as a programmer. One can waste hours tracking down a bug that turns out to be a simple typo, or more likely, an off by one error that plagues much of bioinformatics. I’ve felt the self-directed anger having submitted thousands of cluster jobs that have failed with a missing parameter, or waited hours for a program to complete, only to discover the disk has run out of room to store the output. Yet, these problems pale into comparison in the face of problems at the bench.
current status #microbiology #fuckpcr #fuckyou pic.twitter.com/OgRwI8ueTI
— Sam Nicholls (@samstudio8) September 16, 2017
I’ve spent days in the lab, setting-up and executing PCR, casting, loading and running gels, only to take a UV image of absolutely nothing at all.
BLURRY GARBAGE STRIKES AGAIN pic.twitter.com/n66OwHFhty
— Sam Nicholls (@samstudio8) June 27, 2017
Last year, I spent most of Christmas sheparding data through our cluster, much to my family’s dismay. This year, I had to miss a large family do for a sister’s milestone birthday. I spent many midnights in the lab, lamenting the life of a PhD student, and shuffling around with angry optimism; “Surely it has to fucking work this time?”. Until finally, I got what I wanted.
Fuck yes. Salvaged my weekend. pic.twitter.com/2YxF3qm2AW
— Sam Nicholls (@samstudio8) September 17, 2017
I screamed so loud with glee that security came to check on me. “I’m a fucking scientist now!”
New Nanopore Toys
My experiment was simple in practice. Computationally, I’d predicted haplotypes with my Gretel method from short-read Illumina data from a real rumen microbiome. I designed 10 pairs of primers to capture 10 genes of interest (with hydrolytic-activity) using the haplotypes. And finally, after several weeks of constant almost 24/7 lab work, building cDNA libraries and amplifying the genes of interest, I made enough product for the exciting next step: Nanopore sequencing.
Squee! @nanopore toys of my very own! pic.twitter.com/DusuYisoqq
— Sam Nicholls (@samstudio8) July 28, 2017
With some invaluable assistance from our resident Nanopore expert Arwyn Edwards (@arwynedwards) and PhD student André (@GeoMicroSoares), I sequenced my amplicons on an Oxford Nanopore MinION, and the results were incredible.
I bottled loading the @nanopore myself. @arwynedwards is loading Gretel Mix Mk. 1 and we wait to see if my PhD is ruined. pic.twitter.com/PB76vVSmcb
— Sam Nicholls (@samstudio8) September 22, 2017
Our Nanopore reads strongly supported our haplotypes, and concurred with the Sanger sequencing. Finally, we have empirical biological evidence that Gretel works.
3yrs to define the metahaplome, create data struct & algo to extract it, & do my own wet lab work. 670k reads of proof now live on this disk pic.twitter.com/pQWiPFuC6b
— Sam Nicholls (@samstudio8) September 26, 2017
The pre-print rises
With this bomb-shell in the bag, the third version of my pre-print rose from the ashes of the second. We demoted the DHFR and HIV-1 data sets to the Supplement, and included an analysis on our performance with a de facto benchmark mock community introduced by Chris Quince in its place. The data sets and evaluation mechanisms that our previous reviewers found unrepresentative and convoluted were gone. I even got to include a Circos plot.
paper has a circos plot now so the probability of being accepted for publication has probably increased 10-fold #moneyshot #CIRCLES pic.twitter.com/CHbsb6VCqH
— Sam Nicholls (@samstudio8) November 3, 2017
Once more, we substantially updated the manuscript, and released a new pre-print. We made our to bioRxiv to much Twitter fanfare, earning over 1,500 views in our first week.
It's up! After 18 months of work: a mathematical definition for recovery of enzyme isoforms from metagenomes, a data structure and algorithm for metagenomic haplotype recovery, and finally: biological proof of method with @nanopore https://t.co/yD6pYqstot
— Sam Nicholls (@samstudio8) November 22, 2017
This work also addresses every piece of feedback we’ve had from reviewers in the past. Surely, the publishing process would now finally recognise our work and send us out for review, right?
Desk rejection on a Friday afternoon pic.twitter.com/urOT1iiavQ
— Sam Nicholls (@samstudio8) November 24, 2017
Sadly, the journey of this work is still not smooth sailing, with three of my weekends marred by a Friday desk rejection…
My third Friday evening paper rejection. It's hard enough to maintain a good work-life balance in science as it is without trashing my weekend with news that could have waited until Monday. pic.twitter.com/oT4mripvUT
— Sam Nicholls (@samstudio8) December 8, 2017
…and a fourth desk rejection on the last working day before Christmas was pretty painful. But we are currently grateful to be in discussion with an editor and I am trying to remain hopeful we will get where we want to be in the end. Wish us luck!
In other news…
Of course, I am one for procrastination, and have been keeping busy while all this has been unfolding…
I hosted a national student conference
Hello #symbiosys17!@ISCB_RSG_UK @bcsmidwales @nanopore @repositiveio pic.twitter.com/gmwiUkeeYk
— #Symbiosys17 (@symbiosys17) August 11, 2017
I am applying for some fellowships
Seeing a full grant application form for the first time pic.twitter.com/Bh1NwUMGy6
— Sam Nicholls (@samstudio8) November 9, 2017
I’ve officially started my thesis…
CRITICAL ALERT
THERE IS A DOCUMENT.TEX
I HAVE OFFICIALLY STARTED MY THESIS— Sam Nicholls (@samstudio8) April 9, 2017
…which is just as well, because the money is gone
When your last ever PhD stipend cheque arrives pic.twitter.com/amAZ6RzigX
— Sam Nicholls (@samstudio8) June 30, 2017
I’ve started making cheap lab tat with my best friend…
Time to test out our new @samtomindustrys magnetic separatron! pic.twitter.com/DoLMYNupOE
— Josh Quick (@Scalene) September 4, 2017
…it’s approved by polar bears
PSA: The @samtomindustrys separatron is the only low cost magnetic rack system endorsed by polar bears. Accept no substitutes. pic.twitter.com/tjFpWPuvK0
— Arwyn Edwards (@arwynedwards) December 1, 2017
…and the UK Centre for Astrobiology
.@samtomindustrys Thanks guys! The NASAs loved the sweeties too! pic.twitter.com/ezLAGvnqU8
— AstrobiologyUKCA (@UKAstrobiology) October 9, 2017
…and has been to the Arctic
IT HAS BEEN DRAWN IRRESISTABLY TO THE ULTIMA THULE OF THE NORTH POLE. (Hope to use it in a week or so when things have calmed down a bit.)
— Arwyn Edwards (@arwynedwards) June 30, 2017
I gave an invited talk at a big conference…
It's me! #g10kgs2017 pic.twitter.com/DuAupc8USC
— Sam Nicholls (@samstudio8) August 29, 2017
…it seemed to go down well
Who knew cow rumens were such a hostile place for microbes @samstudio8 #g10kgs2017 pic.twitter.com/axSvJFWP3X
— Benjamen White (@benhwhite) August 30, 2017
I hosted UKIEPC at Aber for the 4th year
All teams now with three correct solutions, amazing work @AberCompSci! But who will be the first to four solutions? #ukiepc pic.twitter.com/B4pS69eCYO
— Sam Nicholls (@samstudio8) October 21, 2017
We’ve applied to fund Monster Lab…
Really excited for our @samtomindustrys @MonsterDNALab public engagement grant application! pic.twitter.com/1tyJCIRWsF
— Sam Nicholls (@samstudio8) December 1, 2017
…and made a website to catalogue our monsters
We sequenced 80 Lego DNA sequences as part of Science Week earlier this year at @AberUni, now the monster genomes have been annotated and made available with phenotypes at https://t.co/IRLQmm4RRi.
Come and see our scary collection! pic.twitter.com/vP7fbkV1kS— Monster Lab (@MonsterDNALab) December 5, 2017
For a change I chose my family over my PhD and had a fucking great Christmas
Nicholls et al. (2017) pic.twitter.com/YzWk94NVuV
— Sam Nicholls (@samstudio8) December 25, 2017
What’s next?
- Get this fucking great paper off my desk and out of my life
- Hopefully get invited to some fellowship interviews
- Continue making cool stuff with Sam and Tom Industrys
- Do more cool stuff with Monster Lab
- Finish this fucking thesis so I can finally do something else
tl;dr
- Happy New Year
- For more information, please re-read